Creating A Dual-Axis Plot using R and ggplot
A brief tutorial on plotting two y-axes in ggplot
- Setting the Scene
- Goal
- Generate the PKPD data
- Creating the Dual Axis Plot
- Automatically defining the scaling function
- Bonus: Styling the Secondary Y axis
Setting the Scene
You have collected data on two variables, both changing with time. The variables are hypothesized to be interconnected, where when one changes, you see a corresponding change in the other. Since this is a PKPD oriented blog, let's imagine our variables are drug concentrations and change in a biomarker (although the techniques in this post can be applied for other types of data).
Goal
We want to generate a plot which shows the change of the two variables over time, highlighting their correlated relationship. Like many other pharmacometricians and data scientists, our plotting weapon-of-choice is ggplot, but unfortunately there aren't any out-of-the-box options to generate a plot with two independent y-axes. Mildly frustrating, but this is by design. Dual y-axes can sometimes be used to generate plots that are confusing (at best) or intentionally misleading (at worst), and the developers of ggplot do not want to encourage undisciplined data visualizations. But... sometimes it makes sense and provides valuable insight into the data (like when we know there is a biologic explanation to the correlation).
Generate the PKPD data
We're going to use mrgsolve to generate some PKPD data from an indirect response model (specifically inhibition of production).
Simulating the data
Take a peek at the dataset structure
| time | CP | RESP |
|---|---|---|
| 0 | 0.0000000 | 40.00000 |
| 1 | 0.7381086 | 38.41351 |
| 2 | 0.9362775 | 36.09367 |
| 3 | 0.9533342 | 34.10246 |
| 4 | 0.9166609 | 32.58596 |
| 5 | 0.8697235 | 31.49649 |
| 6 | 0.8260183 | 30.74640 |
| 7 | 0.7886785 | 30.25182 |
| 8 | 0.7575112 | 29.94408 |
| 9 | 0.7314570 | 29.77050 |
Plot the data
In an initial step to visualize our data, let's just plot the two variables in independent plots and stack them.
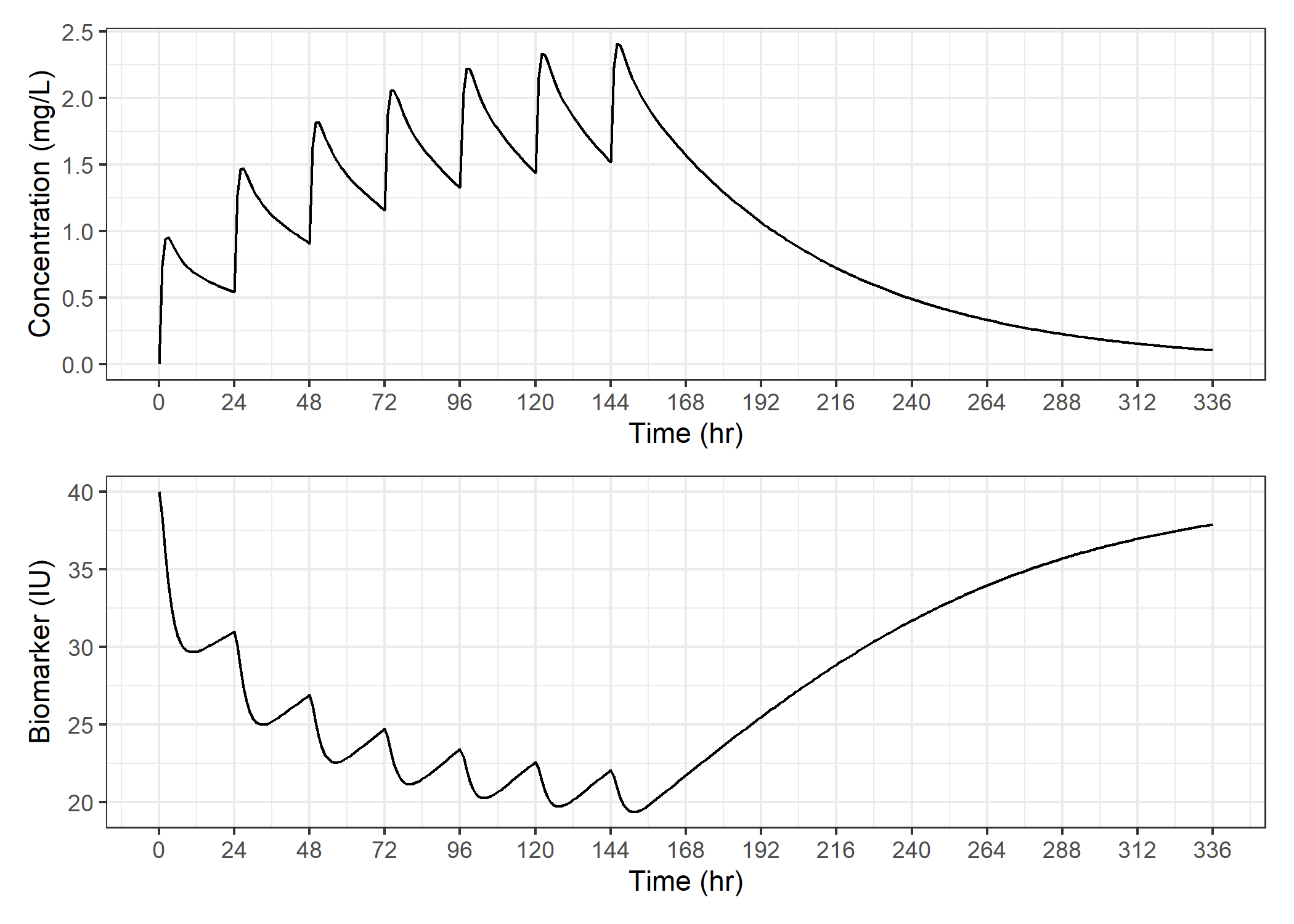
As expected, we see that when concentrations go up, the biomarker decreases due to inhibition of production of the biomarker.
Creating the Dual Axis Plot
Next, we'll make a plot with dual y axes, with the left axis showing the concentration scale and the right axis showing the biomarker scale. Although ggplot doesn't allow creating a separate independent y-axis, it does allow creating a second y axis that is a one-to-one transformation of the first. With some clever manipulation, we can use this feature to create our second axis.
Plotting both series on the same figure
We'll start by plotting both variables on the same figure. We'll modify our PK plot slightly to add the biomarker time series layer.
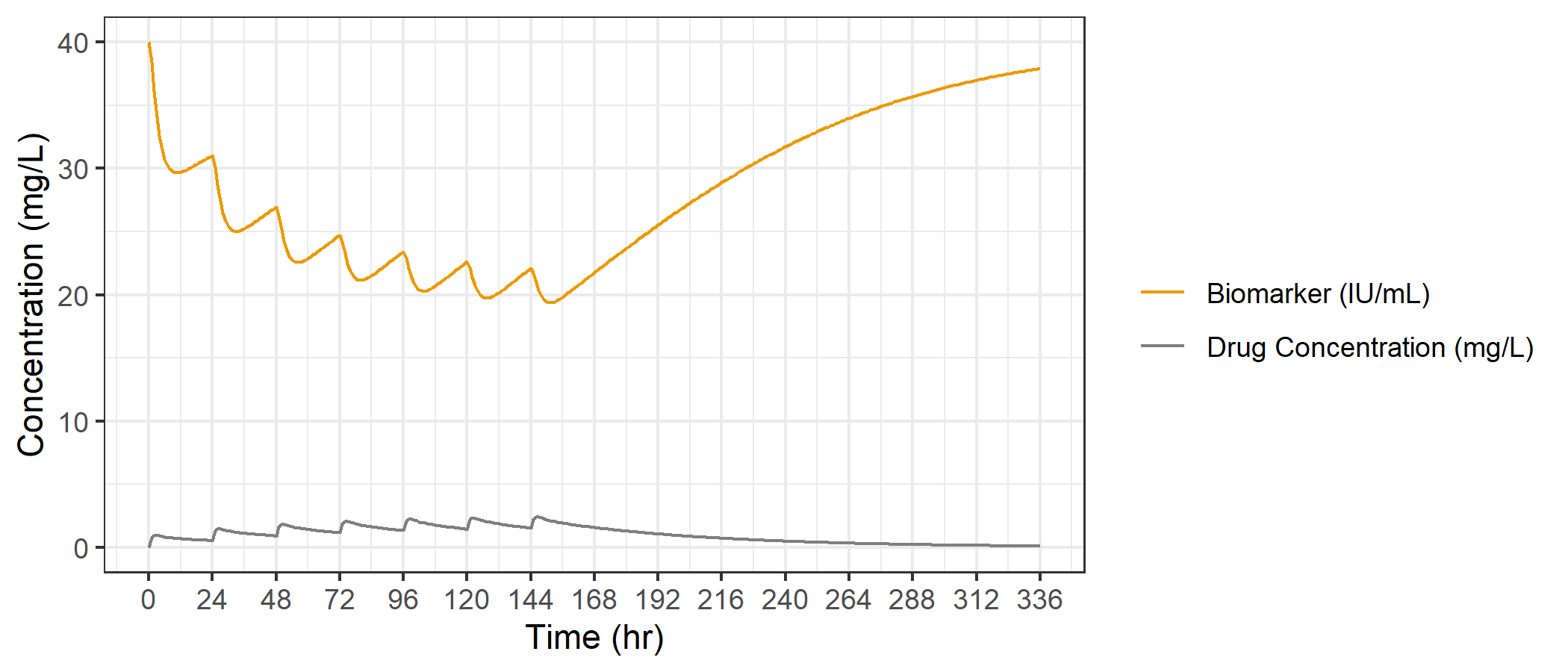
Doesn't look terrible, but since the values of the two variables are so different, there is a lot of empty space in the plot and also the y-axis is only labelled for concentrations.
Adding a second y-axis
To get rid of all the empty space, we need to get the values of the two variables in the same range. Roughly, the range of our response variable is about 15 times higher than the drug concentrations. So, instead of plotting the biomarker response directly, we'll plot it scaled by a factor of 15 (RESP/15). When adding the second y axis, we need to remember to apply that same factor scaling to the axis to maintain the correct values being shown. To add the second axis and scale it by 15, we can include the following line in the ggplot code
To make sure we use the same scaling factor for both the variable transformation and the axis transformation, we'll define a scale variable up front, and use it in both places that we need to.
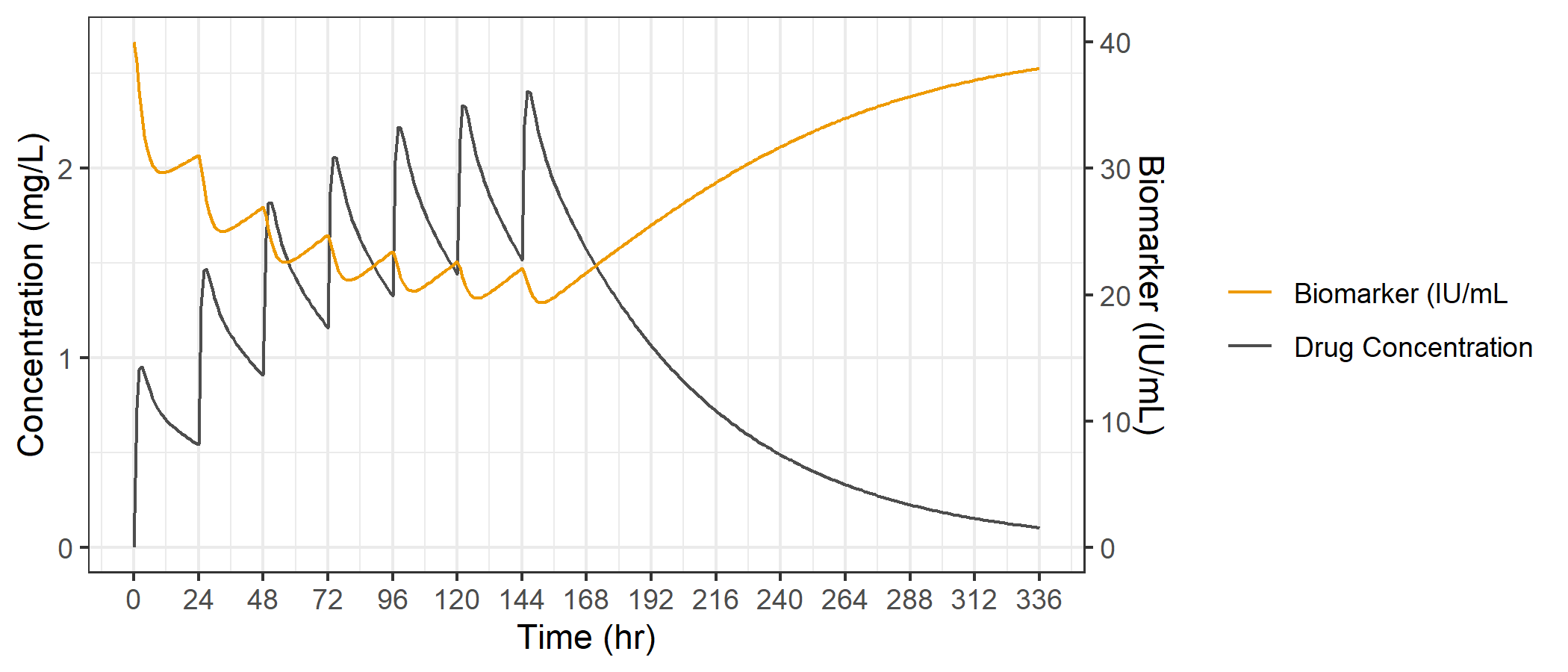
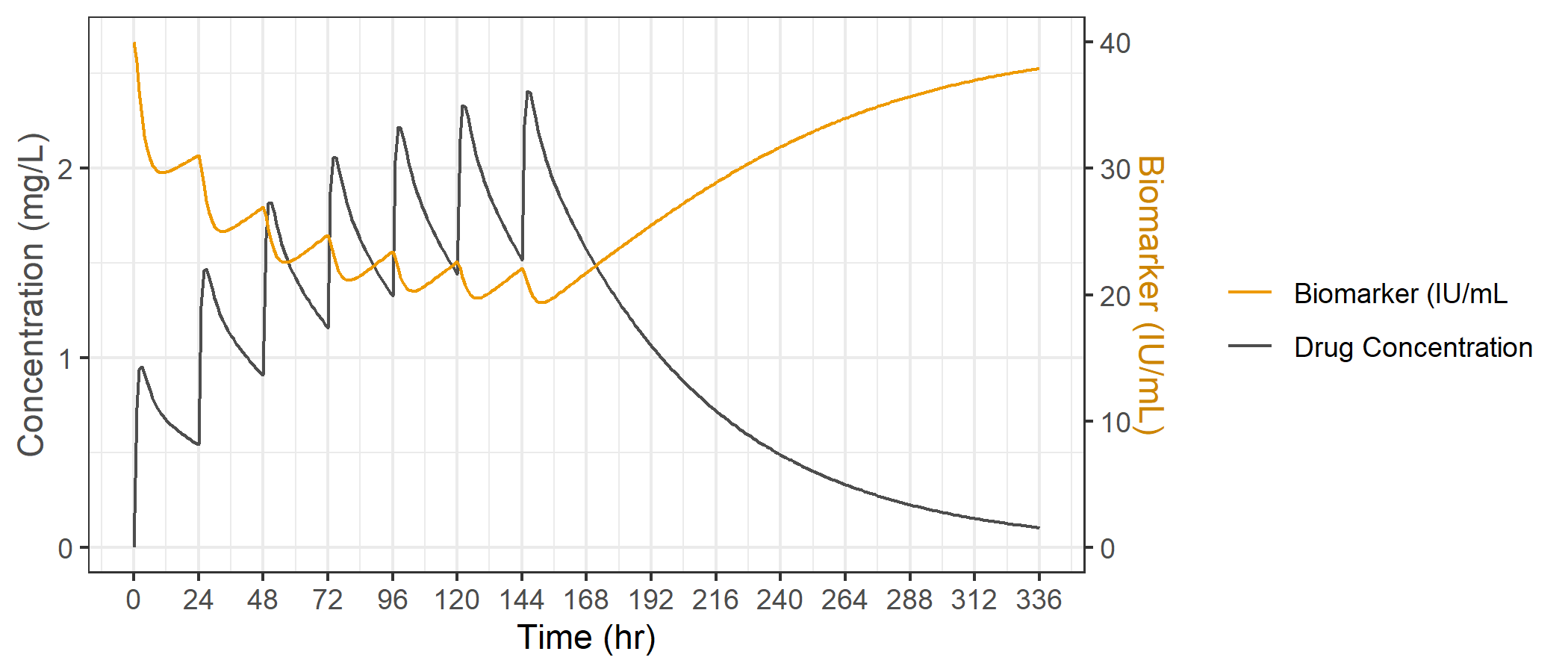
And voila... just like that we have our two variables on the same plot, and we can nicely see an increase in PK concentrations drives a decrease in the biomarker, with a return to baseline upon washout of PK.
Automatically defining the scaling function
Sometimes we'll need a more objective way to choose the scaling function to make the plotting routine more general. A common example is when we have multiple subjects in our dataset and we need to loop through and produce an individual plot for each one. Since drug concentrations and response can vary between subjects, we'll need a smarter way to choose the scaling function based on the individual subject's data.
We'll define the plot axis limits upfront, and use some geometry based transformation fucntions to scale our second variable.

Using this method, we can get perfectly scaled axes every time. If you want to change one of the axis limits (i.e. if you always wanted a lower limit of 0 for both variables) you could just edit the min_first and min_second variables to a value of 0.
Bonus: Styling the Secondary Y axis
We can apply styling to the second Y axis if we want to emphasize which axis corresponds to which variable. Since our drug concentration is colored gray and biomarker is colored orange, we can use those same colors for the axis titles.